By genome sequencing a young child’s tumour, doctors at Addenbrooke's have discovered a new gene mutation which is benign and doesn't requiring cancer treatment.
Shortly after birth, the baby was found to have an unusual tumour which was hard to diagnose, making treatment decisions difficult.
The child was referred to Addenbrooke’s Hospital when their tumour had stabilised, where the clinical team in the first instance decided to monitor how fast it was growing.
Nine months later the team was able to see that tumour had increased in size.
To help choose the best treatment for the child, whole genome sequencing (WGS) was performed through the NHS and results validated through targeted RNA panel testing.
This revealed that the tumour was benign, and also led to the discovery of the new alteration.
This particular child had a very unusual tumour near their wrist. From initial review of the child and of test results from the patient’s former hospital we were not sure what we were dealing with.
Dr Sam Behjati, Paediatric Oncologist, CUH and group leader at the Wellcome Sanger Institute
Dr Behjati is a paediatric oncologist at Cambridge University Hospitals NHS Foundation Trust (CUH) and group leader at the Wellcome Sanger Institute. He said:
“When the tumour started to grow, we were concerned that it could be an aggressive cancer requiring cancer treatment, including aggressive surgery, which would have put the child at risk of not being able to use their hand fully in the future.”
WGS is available to all children being treated in the NHS with confirmed or suspected cancer through the NHS National Genomic Medicine Service and Test Directory.
At CUH, paediatric oncologists work closely with scientists in NHS East Genomic Laboratory Hub to provide this very useful test to all such patients as routine practice with patient or family consent.
In this case, WGS alongside other tests helped confirm a rare and unexpected diagnosis – desmoplastic fibroblastoma – which is not cancerous. The patient therefore did not need to go through major surgery or aggressive treatment, which was great news for the family.
Dr Behjati




Genomic tests and validation of results were carried out through NHS East Genomic Laboratory Hub and NHS North Thames Genomic Laboratory Hub (GLH).
Genomics England partner with the NHS to provide whole genome sequencing diagnostics; the data and findings generated by Genomics England are sent to the NHS Laboratory team for interpretation and reporting to the clinical team.
This case highlights the utility of WGS for discovering the molecular causes of suspected cancer in children.
Professor Mike Hubank, Scientific Lead of the North Thames GLH and Professor of Translational Genomics at The Institute of Cancer Research, London
Professor Mike Hubank added:
"It also demonstrates how the NHS Genomic Medicine Service in England is world leading in providing a state-of-the-art toolkit of genomic cancer tests to all patients, free at the point of care and tailored to their individual needs. In this case, collaboration across regional laboratories helped verify that the growth was not cancerous.
“The tailored, cost-effective application of genomic testing within the NHS will improve cancer diagnosis and more accurately match cancer treatments with the patients who are most likely to benefit.”

This type of tumour is quite rare, and not one we would expect to see in children. “Whole genome sequencing showed a specific rearrangement of the FOS Like 1 (FOSL1) gene, which has not been seen in human tumours to date.
Dr Solange De Noon, Clinical Research Fellow, University College London Cancer Institute and Wellcome Sanger Institute,
Dr Solange added:
“Using the Royal National Orthopaedic Hospital (RNOH) biobank, we compared the findings to those of 15 other cases with the same diagnosis – all adult cases - and this rearrangement was also present in these cases.
“This rearrangement will help clarify the type of tumour in many future cases. Furthermore, it was not demonstrated in a tumour with many similar features seen down the microscope (fibroma of tendon sheath) making it a useful diagnostic tool.
“This finding confirms that genomic studies through the NHS Genomic Medicine Service will lead to new discoveries.”
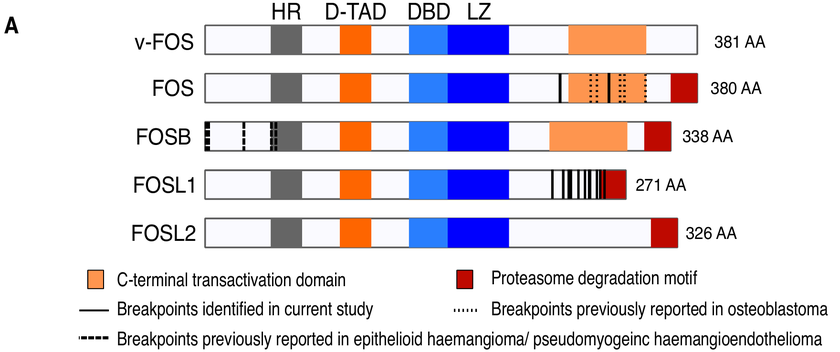
The FOSL1 gene belongs to the FOS gene family (FOS, FOSB, FOSL1, and FOSL2) which are known to be altered in rare bone and soft tissue tumours.
Previous studies have identified high expression of the FOSL1 gene in this uncommon benign soft-tissue tumour known as ‘desmoplastic fibroblastoma’, but have failed to pinpoint the underlying cause.

This is another example of how genomics can help deliver more accurate diagnoses for cancer patients and more personalised and targeted treatment.
Professor Dame Sue Hill, Chief Scientific Officer for England and Senior Responsible Officer for Genomics in NHS England
Professor Dame Sue Hill is Chief Scientific Officer for England and Senior Responsible Officer for Genomics in NHS England. She said:
“This testing is available to all children with cancer and, through the NHS’ Genomics Strategy which sets out our vision for embedding genomics in the NHS over the next five years, more patients and their families will be able to benefit.”
The paper has been published in the Journal of Pathology. (opens in a new tab)
This study was funded by Wellcome, The Tom Prince Cancer Trust, Sarcoma UK the Bone Cancer Research Trust and the Royal National Orthopaedic Hospital NHS Research and Development grant. Full detail is included within the publication.
